Background
Head and neck cancer is the sixth leading cause of cancer worldwide. The 5-year survival rate is poor, at an estimated 50% overall. Current treatment modalities cause significant long-term morbidity which greatly impacts quality of life of survivors. Therefore, there is an urgent need to develop novel therapeutic strategies that would decrease recurrence rate and treatment related-toxicity and improve clinical outcomes and overall survival. Our lab is focusing on head and neck squamous cell carcinoma (HNSCC) but is also examining other head and neck malignancies. We aim to find innovative therapeutic approaches and diagnostic methods to better overcome challenges encountered in clinical practice. These approaches will be based on thorough and comprehensive investigation of biologic and molecular processes in head and neck cancer. To perform this task we are using a wide range of tissue cultures, primary cells (human and mouse) as well as animal models. Our tools span a wide range from classic molecular biology to cutting edge tools and methods such as next generation sequencing.
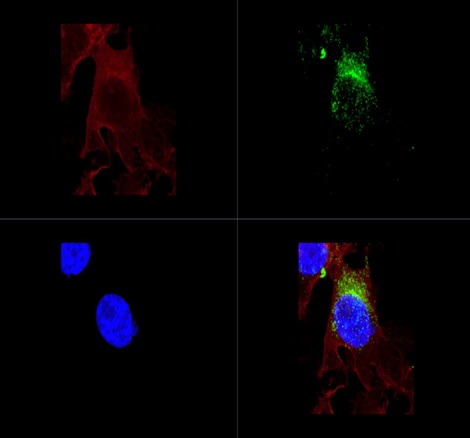
Figure1. Labelled exosomes. Top left – phalloidin marking the actin cellular structure;
Top right – exosomes marked using CD63-GFP;
Bottom left – Dapi staining of cellular nuclei; Bottom right – combined image
Currently, we concentrate our efforts on the HNSCC, its interaction with supporting fibroblast cells and environment in general. The p53 tumor suppressor gene plays a critical role in HNSCC. It is regulating key cellular pathways including apoptosis and cell cycle control. In cases where p53 is mutated, it not only loses its tumor suppressive functions, but also gains novel tumorigenic functions thus acting as an oncogene. RNA interference is a native cellular mechanism responsible for regulating gene expression. It has been widely shown that small interfering RNAs (siRNA) can be utilized to knockdown specific oncogenes resulting in the deliberate inhibition of cancer propagation. The siRNA can be delivered more efficiently to cancer cells and tumors either via liposomes which are synthetic micelles or via exosomes which are produced by cells naturally
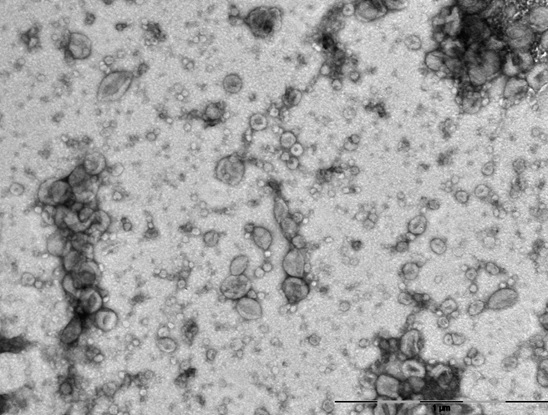
Figure2. TEM scanned exosomes.
These exosomes were harvested from mouse squamous cell carcinoma.
Not only are the tumor cells of interest, but also their interaction with the immediate environment. Cancer cells require a multifactorial support system for them to develop into life-threatening tumors and metastasize to distant locations. In order for cancer cells to survive and proliferate, they must be supported by their microenvironment, including the extracellular matrix, blood vessels, growth factors, and fibroblasts. It has been shown that fibroblasts in the tumor microenvironment are phenotypically and functionally different from normal fibroblasts, and are called cancer associated fibroblasts (CAFs) or myofibroblasts, because of their contraction properties. They express pro-inflammatory genes (Erez, Glanz, Raz, Avivi, & Barshack, 2013) and promote tumor growth by enhancing tumor cell proliferation and angiogenesis, and also contribute to drug resistance of cancer cells. Fibroblasts activation is a consequence of a variety of factors, which are secreted by cancer cells, such as TGF-β, HGF, PDGF and FGF. Moreover, cancer cells can activate fibroblasts via exosomal miRNAs (Yang et al., 2017). Previous studies testing CAFs from head and neck cancer tissues have identified miRNAs overexpressed in CAFs compared with their paired normal fibroblasts utilizing microRNA array analyses, such as mir-7.
In order to find new strategies to induce tumor growth inhibition, understanding the communication between head and neck cancer cells and the tumor microenvironment components is vital
Cancer research also involves early detection and post-operative surveillance. Circulating DNA (circDNA) is DNA that flows in the blood normally. Cancer cells also release DNA into the blood stream when they go through apoptosis. Detection of tumor-originated circulating DNA is feasible using highly sensitive Digital PCR amplification protocols and equipment. Developing these molecular tools will enable early cancer detection in healthy subjects before initial signs are noticed. It will also become an important tool monitoring patients for cancer recurrence after potentially curative therapy.
Research Topics
- We are examining cancer-cells targeting and siRNA-based gene silencing. This is done via exosomes and lipid-based nanpoparticles (LNPs) as endogenous and synthetic (respectively) delivery systems.
- We are interested in targeting nanovasicles (exosomes or LNPs) toward cancer cells for therapeutic, diagnostics and other applications.
- We are characterizing the cancer environment, more specifically CAF's, in order to examine the relationship between cancer cells and fibroblasts, for the prevention of key steps in cancer growth.
- The lab is using cutting edge molecular tools and technologies to examine the genetic signature of recurrent disease.
- We are designing novel molecular tools to detect cancer circDNA in blood samples ("liquid biopsy") or other body fluids before any physical signs appear. These tools will also be used to monitor cancer recurrence or metastases.
Laboratory members
- Saar Tal, PhD
- Moran Penn, Msc. MD-PhD student
- Liyona Kampel, MD. PhD student
Collaborations:
- Dr. Dov Hershkovitz, Ichilov Medical center, Department of Pathology
- Prof. Neta Erez, Tel Aviv University, Department of Pathology
- Prof. Dan Peer, Tel Aviv University, Department of Cell Research & Immunology
- Prof. Zelig Eshchar, Weizmann institute, Department of Immunology
- Dr. Noam Shomron, Tel Aviv University, Department of Cell and Developmental Biology
Contact us
Current laboratory Funding
Organization Total Amount/year Duration
ISF 185,000 NIS 3 years
Alrov Fundation 75,000 1 year
References
1.Diehl, F., Schmidt, K., Choti, M. A., Romans, K., Goodman, S., Li, M., … Jr, L. A. D. (2008). Circulating mutant DNA to assess tumor dynamics, 14(9), 985–990. https://doi.org/10.1038/nm.1789
2.Efrati, E., Elkin, H., Peerless, Y., Sabo, E., Ben-Izhak, O., Hershkovitz, D. (2011) Cancer bFiomarkers. 8: 89-94. doi: 10.3233/CBM-2011-0203
3.Gale, D., Sc, B., Forshew, T., Ph, D., Mahler-araujo, B., Rajan, S., … Sci, F. M. (2013). Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer, 1199–1209. https://doi.org/10.1056/NEJMoa1213261
4.Gatta, G., Botta, L., Sánchez, M. J., Anderson, L. A., Pierannunzio, D., & Licitra, L. (2015). Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. European Journal of Cancer, 51(15), 2130–2143. https://doi.org/10.1016/j.ejca.2015.07.043
5.Guo, N., Lou, F., Ma, Y., Li, J., Yang, B., Chen, W., … Jones, L. (2016). Circulating tumor DNA detection in lung cancer patients before and after surgery. Nature Publishing Group, (September), 1–8. https://doi.org/10.1038/srep33519
6.Lee, N., Xia, P., Fischbein, N. J., Akazawa, P., Akazawa, C., & Quivey, J. M. (2003). Intensity-modulated radiation therapy for head-and-neck cancer: The UCSF experience focusing on target volume delineation. International Journal of Radiation Oncology Biology Physics, 57(1), 49–60. https://doi.org/10.1016/S0360-3016(03)00405-X
7.Muhanna, N., Grappa, M., Chan, H. Bratman, S.V. (2017) Cell free DNA kinetics in a pre-clinical model of head and neck caner. Nature Scientific Reports. 7(1):16723. DOI: 10.1038/s41598-017-17079-6
8.Perets, R., Greenberg, O., Shentzer, T….Hershkovitz, D. (2018) Oncologist. 23 (5): 566-572. doi: 10.1634/theoncologist.2017-0467
9.POSTEL, M., ROOSEN, A., LAURENT-PUIG, P., TALY, V., & WANG-RENAULT, S.-F. (2017). Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Review of Molecular Diagnostics, 14737159.2018.1400384. https://doi.org/10.1080/14737159.2018.1400384
10.Siegel, R., Miller, K. D., & Ahmedin, J. (2017). Cáncer Statistics. Ca Cáncer Journal, 67(1), 7–30. https://doi.org/10.3322/caac.21387.
11.Stransky, N., Egloff, A. M., Tward, A. D., Kostic, A. D., Cibulskis, K., Sivachenko, A., … Grandis, J. R. (2011). The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science, 333(6046), 1157–1160. https://doi.org/10.1126/science.1208130
12.TCGA Network. (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 517(7536), 576–582. https://doi.org/10.1038/nature14129
13.Van Ginkel, J. H., Huibers, M. M. H., Noorlag, R., De Bree, R., Van Es, R. J. J., & Willems, S. M. (2017). Liquid biopsy: A future tool for posttreatment surveillance in head and neck cancer? Pathobiology, 84(3), 115–120. https://doi.org/10.1159/000452861
14.Volik, S., Alcaide, M., Morin, R. D., & Collins, C. (2016). Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Molecular Cancer Research, 14(10), 898–908. https://doi.org/10.1158/1541-7786.MCR-16-0044
15.Yang, H., Wang, K., Angeles, L., Angeles, L., Angeles, L., & Angeles, L. (2016). HHS Public Access, 10(10), 1556–1566.
16.Erez, N., Glanz, S., Raz, Y., Avivi, C., & Barshack, I. (2013). Cancer Associated Fibroblasts express pro-inflammatory factors in human breast and ovarian tumors. Biochemical and Biophysical Research Communications, 437(3), 397–402. https://doi.org/https://doi.org/10.1016/j.bbrc.2013.06.089
17.Paraiso, K. H. T., & Smalley, K. S. M. (2013). Fibroblast-mediated drug resistance in cancer. Biochemical Pharmacology, 85(8), 1033–1041. https://doi.org/https://doi.org/10.1016/j.bcp.2013.01.018
18.Shen, Z., Qin, X., Yan, M., Li, R., Chen, G., Zhang, J., & Chen, W. (2017). Cancer-associated fibroblasts promote cancer cell growth through a miR-7-RASSF2-PAR-4 axis in the tumor microenvironment. Oncotarget, 8(1), 1290–1303. https://doi.org/10.18632/oncotarget.13609
19.Shiga, K., Hara, M., Nagasaki, T., Sato, T., Takahashi, H., & Takeyama, H. (2015). Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers, 7(4), 2443–2458. https://doi.org/10.3390/cancers7040902
20.Yang, F., Ning, Z., Ma, L., Liu, W., Shao, C., Shu, Y., & Shen, H. (2017). Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Molecular Cancer, 16(1), 148. https://doi.org/10.1186/s12943-017-0718-4